 在 TensorFlow.org 上檢視 在 TensorFlow.org 上檢視
|
 在 Google Colab 中執行 在 Google Colab 中執行
|
 在 GitHub 上檢視原始碼 在 GitHub 上檢視原始碼
|
 下載筆記本 下載筆記本
|
在這個 Colab 中,我們將探索 TensorFlow Probability 的一些基本功能。
依附元件與先決條件
匯入
公用程式
大綱
- TensorFlow
- TensorFlow Probability
- 分佈
- 雙射器
- MCMC
- ...以及更多!
前言:TensorFlow
TensorFlow 是一個科學運算程式庫。
它支援
- 大量的數學運算
- 有效率的向量化運算
- 輕鬆的硬體加速
- 自動微分
向量化
- 向量化讓速度更快!
- 這也表示我們經常思考形狀
mats = tf.random.uniform(shape=[1000, 10, 10])
vecs = tf.random.uniform(shape=[1000, 10, 1])
def for_loop_solve():
return np.array(
[tf.linalg.solve(mats[i, ...], vecs[i, ...]) for i in range(1000)])
def vectorized_solve():
return tf.linalg.solve(mats, vecs)
# Vectorization for the win!
%timeit for_loop_solve()
%timeit vectorized_solve()
1 loops, best of 3: 2 s per loop 1000 loops, best of 3: 653 µs per loop
硬體加速
# Code can run seamlessly on a GPU, just change Colab runtime type
# in the 'Runtime' menu.
if tf.test.gpu_device_name() == '/device:GPU:0':
print("Using a GPU")
else:
print("Using a CPU")
Using a CPU
自動微分
a = tf.constant(np.pi)
b = tf.constant(np.e)
with tf.GradientTape() as tape:
tape.watch([a, b])
c = .5 * (a**2 + b**2)
grads = tape.gradient(c, [a, b])
print(grads[0])
print(grads[1])
tf.Tensor(3.1415927, shape=(), dtype=float32) tf.Tensor(2.7182817, shape=(), dtype=float32)
TensorFlow Probability
TensorFlow Probability 是一個用於 TensorFlow 中機率推理和統計分析的程式庫。
我們透過組合低階模組化元件來支援建模、推論和評估。
低階建構區塊
- 分佈
- 雙射器
高 (較高) 階層的建構
- 馬可夫鏈蒙地卡羅
- 機率層
- 結構性時間序列
- 廣義線性模型
- 最佳化工具
分佈
tfp.distributions.Distribution 是一個具有兩個核心方法的類別:sample 和 log_prob。
TFP 有很多分佈!
print_subclasses_from_module(tfp.distributions, tfp.distributions.Distribution)
Autoregressive, BatchReshape, Bates, Bernoulli, Beta, BetaBinomial, Binomial Blockwise, Categorical, Cauchy, Chi, Chi2, CholeskyLKJ, ContinuousBernoulli Deterministic, Dirichlet, DirichletMultinomial, Distribution, DoublesidedMaxwell Empirical, ExpGamma, ExpRelaxedOneHotCategorical, Exponential, FiniteDiscrete Gamma, GammaGamma, GaussianProcess, GaussianProcessRegressionModel GeneralizedNormal, GeneralizedPareto, Geometric, Gumbel, HalfCauchy, HalfNormal HalfStudentT, HiddenMarkovModel, Horseshoe, Independent, InverseGamma InverseGaussian, JohnsonSU, JointDistribution, JointDistributionCoroutine JointDistributionCoroutineAutoBatched, JointDistributionNamed JointDistributionNamedAutoBatched, JointDistributionSequential JointDistributionSequentialAutoBatched, Kumaraswamy, LKJ, Laplace LinearGaussianStateSpaceModel, LogLogistic, LogNormal, Logistic, LogitNormal Mixture, MixtureSameFamily, Moyal, Multinomial, MultivariateNormalDiag MultivariateNormalDiagPlusLowRank, MultivariateNormalFullCovariance MultivariateNormalLinearOperator, MultivariateNormalTriL MultivariateStudentTLinearOperator, NegativeBinomial, Normal, OneHotCategorical OrderedLogistic, PERT, Pareto, PixelCNN, PlackettLuce, Poisson PoissonLogNormalQuadratureCompound, PowerSpherical, ProbitBernoulli QuantizedDistribution, RelaxedBernoulli, RelaxedOneHotCategorical, Sample SinhArcsinh, SphericalUniform, StudentT, StudentTProcess TransformedDistribution, Triangular, TruncatedCauchy, TruncatedNormal, Uniform VariationalGaussianProcess, VectorDeterministic, VonMises VonMisesFisher, Weibull, WishartLinearOperator, WishartTriL, Zipf
簡單的純量變數 Distribution
# A standard normal
normal = tfd.Normal(loc=0., scale=1.)
print(normal)
tfp.distributions.Normal("Normal", batch_shape=[], event_shape=[], dtype=float32)
# Plot 1000 samples from a standard normal
samples = normal.sample(1000)
sns.distplot(samples)
plt.title("Samples from a standard Normal")
plt.show()
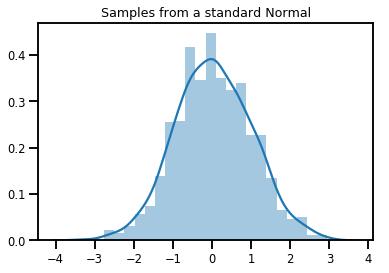
# Compute the log_prob of a point in the event space of `normal`
normal.log_prob(0.)
<tf.Tensor: shape=(), dtype=float32, numpy=-0.9189385>
# Compute the log_prob of a few points
normal.log_prob([-1., 0., 1.])
<tf.Tensor: shape=(3,), dtype=float32, numpy=array([-1.4189385, -0.9189385, -1.4189385], dtype=float32)>
分佈與形狀
Numpy ndarrays 和 TensorFlow Tensors 具有形狀。
TensorFlow Probability Distributions 具有形狀語意 - 我們將形狀劃分為語意上不同的部分,即使相同的記憶體區塊 (Tensor/ndarray) 用於整個所有項目。
- 批次形狀表示具有不同參數的
Distribution集合 - 事件形狀表示來自
Distribution的樣本形狀。
我們總是將批次形狀放在「左邊」,事件形狀放在「右邊」。
純量變數 Distributions 的批次
批次就像「向量化」分佈:獨立執行個體,其運算平行發生。
# Create a batch of 3 normals, and plot 1000 samples from each
normals = tfd.Normal([-2.5, 0., 2.5], 1.) # The scale parameter broadacasts!
print("Batch shape:", normals.batch_shape)
print("Event shape:", normals.event_shape)
Batch shape: (3,) Event shape: ()
# Samples' shapes go on the left!
samples = normals.sample(1000)
print("Shape of samples:", samples.shape)
Shape of samples: (1000, 3)
# Sample shapes can themselves be more complicated
print("Shape of samples:", normals.sample([10, 10, 10]).shape)
Shape of samples: (10, 10, 10, 3)
# A batch of normals gives a batch of log_probs.
print(normals.log_prob([-2.5, 0., 2.5]))
tf.Tensor([-0.9189385 -0.9189385 -0.9189385], shape=(3,), dtype=float32)
# The computation broadcasts, so a batch of normals applied to a scalar
# also gives a batch of log_probs.
print(normals.log_prob(0.))
tf.Tensor([-4.0439386 -0.9189385 -4.0439386], shape=(3,), dtype=float32)
# Normal numpy-like broadcasting rules apply!
xs = np.linspace(-6, 6, 200)
try:
normals.log_prob(xs)
except Exception as e:
print("TFP error:", e.message)
TFP error: Incompatible shapes: [200] vs. [3] [Op:SquaredDifference]
# That fails for the same reason this does:
try:
np.zeros(200) + np.zeros(3)
except Exception as e:
print("Numpy error:", e)
Numpy error: operands could not be broadcast together with shapes (200,) (3,)
# But this would work:
a = np.zeros([200, 1]) + np.zeros(3)
print("Broadcast shape:", a.shape)
Broadcast shape: (200, 3)
# And so will this!
xs = np.linspace(-6, 6, 200)[..., np.newaxis]
# => shape = [200, 1]
lps = normals.log_prob(xs)
print("Broadcast log_prob shape:", lps.shape)
Broadcast log_prob shape: (200, 3)
# Summarizing visually
for i in range(3):
sns.distplot(samples[:, i], kde=False, norm_hist=True)
plt.plot(np.tile(xs, 3), normals.prob(xs), c='k', alpha=.5)
plt.title("Samples from 3 Normals, and their PDF's")
plt.show()
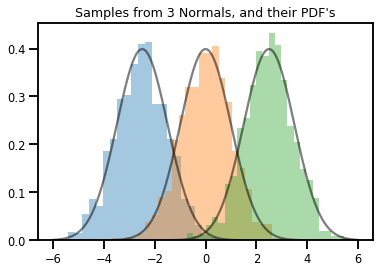
向量變數 Distribution
mvn = tfd.MultivariateNormalDiag(loc=[0., 0.], scale_diag = [1., 1.])
print("Batch shape:", mvn.batch_shape)
print("Event shape:", mvn.event_shape)
Batch shape: () Event shape: (2,)
samples = mvn.sample(1000)
print("Samples shape:", samples.shape)
Samples shape: (1000, 2)
g = sns.jointplot(x=samples[:, 0], y=samples[:, 1], kind='scatter')
plt.show()
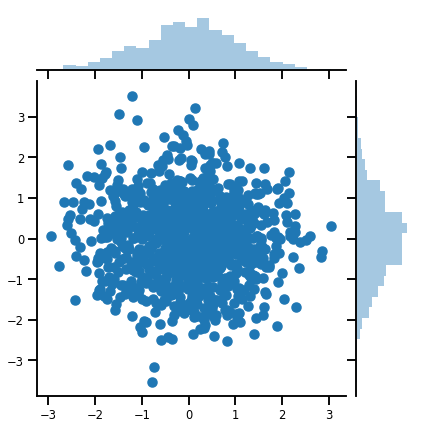
矩陣變數 Distribution
lkj = tfd.LKJ(dimension=10, concentration=[1.5, 3.0])
print("Batch shape: ", lkj.batch_shape)
print("Event shape: ", lkj.event_shape)
Batch shape: (2,) Event shape: (10, 10)
samples = lkj.sample()
print("Samples shape: ", samples.shape)
Samples shape: (2, 10, 10)
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(6, 3))
sns.heatmap(samples[0, ...], ax=axes[0], cbar=False)
sns.heatmap(samples[1, ...], ax=axes[1], cbar=False)
fig.tight_layout()
plt.show()
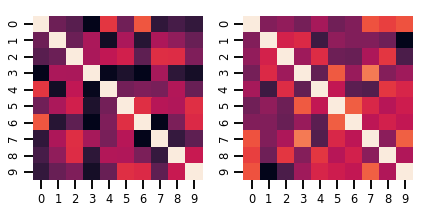
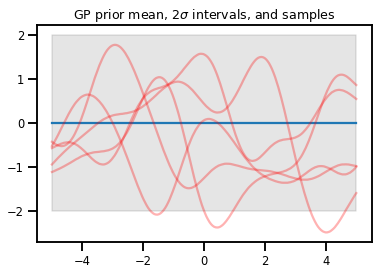
高斯過程
kernel = tfp.math.psd_kernels.ExponentiatedQuadratic()
xs = np.linspace(-5., 5., 200).reshape([-1, 1])
gp = tfd.GaussianProcess(kernel, index_points=xs)
print("Batch shape:", gp.batch_shape)
print("Event shape:", gp.event_shape)
Batch shape: () Event shape: (200,)
upper, lower = gp.mean() + [2 * gp.stddev(), -2 * gp.stddev()]
plt.plot(xs, gp.mean())
plt.fill_between(xs[..., 0], upper, lower, color='k', alpha=.1)
for _ in range(5):
plt.plot(xs, gp.sample(), c='r', alpha=.3)
plt.title(r"GP prior mean, $2\sigma$ intervals, and samples")
plt.show()
# *** Bonus question ***
# Why do so many of these functions lie outside the 95% intervals?
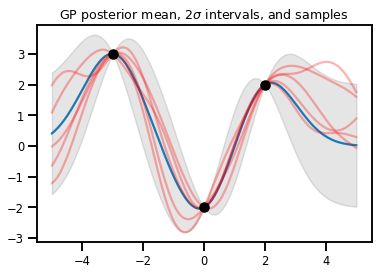
GP 迴歸
# Suppose we have some observed data
obs_x = [[-3.], [0.], [2.]] # Shape 3x1 (3 1-D vectors)
obs_y = [3., -2., 2.] # Shape 3 (3 scalars)
gprm = tfd.GaussianProcessRegressionModel(kernel, xs, obs_x, obs_y)
upper, lower = gprm.mean() + [2 * gprm.stddev(), -2 * gprm.stddev()]
plt.plot(xs, gprm.mean())
plt.fill_between(xs[..., 0], upper, lower, color='k', alpha=.1)
for _ in range(5):
plt.plot(xs, gprm.sample(), c='r', alpha=.3)
plt.scatter(obs_x, obs_y, c='k', zorder=3)
plt.title(r"GP posterior mean, $2\sigma$ intervals, and samples")
plt.show()
雙射器
雙射器代表 (大部分) 可逆、平滑的函數。它們可用於轉換分佈,保留取得樣本和計算 log_probs 的能力。它們可以在 tfp.bijectors 模組中找到。
每個雙射器至少實作 3 個方法
forward,inverse,以及- 至少
forward_log_det_jacobian和inverse_log_det_jacobian其中之一。
有了這些要素,我們可以轉換分佈,並且仍然從結果中取得樣本和對數機率!
在數學中,有點草率
- \(X\) 是具有 pdf \(p(x)\) 的隨機變數
- \(g\) 是 \(X\) 空間上的平滑可逆函數
- \(Y = g(X)\) 是一個新的轉換隨機變數
- \(p(Y=y) = p(X=g^{-1}(y)) \cdot |\nabla g^{-1}(y)|\)
快取
雙射器也會快取 forward 和 inverse 運算,以及 log-det-Jacobians,這讓我們可以節省重複可能非常昂貴的運算!
print_subclasses_from_module(tfp.bijectors, tfp.bijectors.Bijector)
AbsoluteValue, Affine, AffineLinearOperator, AffineScalar, BatchNormalization Bijector, Blockwise, Chain, CholeskyOuterProduct, CholeskyToInvCholesky CorrelationCholesky, Cumsum, DiscreteCosineTransform, Exp, Expm1, FFJORD FillScaleTriL, FillTriangular, FrechetCDF, GeneralizedExtremeValueCDF GeneralizedPareto, GompertzCDF, GumbelCDF, Identity, Inline, Invert IteratedSigmoidCentered, KumaraswamyCDF, LambertWTail, Log, Log1p MaskedAutoregressiveFlow, MatrixInverseTriL, MatvecLU, MoyalCDF, NormalCDF Ordered, Pad, Permute, PowerTransform, RationalQuadraticSpline, RayleighCDF RealNVP, Reciprocal, Reshape, Scale, ScaleMatvecDiag, ScaleMatvecLU ScaleMatvecLinearOperator, ScaleMatvecTriL, ScaleTriL, Shift, ShiftedGompertzCDF Sigmoid, Sinh, SinhArcsinh, SoftClip, Softfloor, SoftmaxCentered, Softplus Softsign, Split, Square, Tanh, TransformDiagonal, Transpose, WeibullCDF
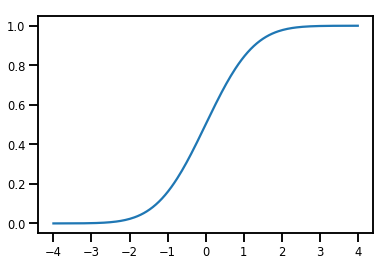
簡單的 Bijector
normal_cdf = tfp.bijectors.NormalCDF()
xs = np.linspace(-4., 4., 200)
plt.plot(xs, normal_cdf.forward(xs))
plt.show()
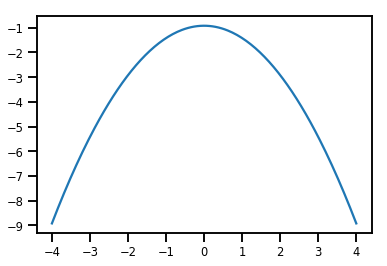
plt.plot(xs, normal_cdf.forward_log_det_jacobian(xs, event_ndims=0))
plt.show()
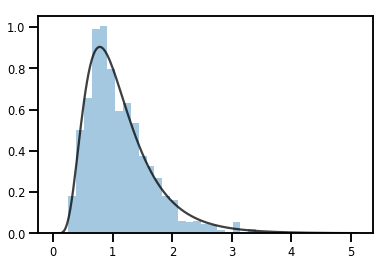
轉換 Distribution 的 Bijector
exp_bijector = tfp.bijectors.Exp()
log_normal = exp_bijector(tfd.Normal(0., .5))
samples = log_normal.sample(1000)
xs = np.linspace(1e-10, np.max(samples), 200)
sns.distplot(samples, norm_hist=True, kde=False)
plt.plot(xs, log_normal.prob(xs), c='k', alpha=.75)
plt.show()
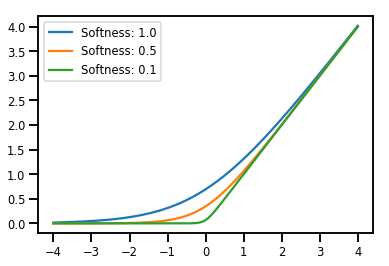
批次處理 Bijectors
# Create a batch of bijectors of shape [3,]
softplus = tfp.bijectors.Softplus(
hinge_softness=[1., .5, .1])
print("Hinge softness shape:", softplus.hinge_softness.shape)
Hinge softness shape: (3,)
# For broadcasting, we want this to be shape [200, 1]
xs = np.linspace(-4., 4., 200)[..., np.newaxis]
ys = softplus.forward(xs)
print("Forward shape:", ys.shape)
Forward shape: (200, 3)
# Visualization
lines = plt.plot(np.tile(xs, 3), ys)
for line, hs in zip(lines, softplus.hinge_softness):
line.set_label("Softness: %1.1f" % hs)
plt.legend()
plt.show()
快取
# This bijector represents a matrix outer product on the forward pass,
# and a cholesky decomposition on the inverse pass. The latter costs O(N^3)!
bij = tfb.CholeskyOuterProduct()
size = 2500
# Make a big, lower-triangular matrix
big_lower_triangular = tf.eye(size)
# Squaring it gives us a positive-definite matrix
big_positive_definite = bij.forward(big_lower_triangular)
# Caching for the win!
%timeit bij.inverse(big_positive_definite)
%timeit tf.linalg.cholesky(big_positive_definite)
10000 loops, best of 3: 114 µs per loop 1 loops, best of 3: 208 ms per loop
MCMC
TFP 內建支援一些標準的馬可夫鏈蒙地卡羅演算法,包括哈密頓蒙地卡羅。
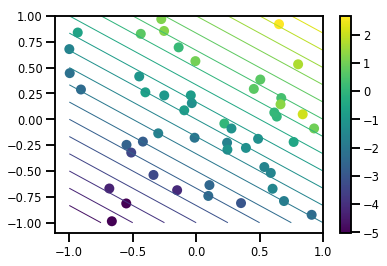
產生資料集
# Generate some data
def f(x, w):
# Pad x with 1's so we can add bias via matmul
x = tf.pad(x, [[1, 0], [0, 0]], constant_values=1)
linop = tf.linalg.LinearOperatorFullMatrix(w[..., np.newaxis])
result = linop.matmul(x, adjoint=True)
return result[..., 0, :]
num_features = 2
num_examples = 50
noise_scale = .5
true_w = np.array([-1., 2., 3.])
xs = np.random.uniform(-1., 1., [num_features, num_examples])
ys = f(xs, true_w) + np.random.normal(0., noise_scale, size=num_examples)
# Visualize the data set
plt.scatter(*xs, c=ys, s=100, linewidths=0)
grid = np.meshgrid(*([np.linspace(-1, 1, 100)] * 2))
xs_grid = np.stack(grid, axis=0)
fs_grid = f(xs_grid.reshape([num_features, -1]), true_w)
fs_grid = np.reshape(fs_grid, [100, 100])
plt.colorbar()
plt.contour(xs_grid[0, ...], xs_grid[1, ...], fs_grid, 20, linewidths=1)
plt.show()
定義我們的聯合對數機率函數
未正規化的後驗是封閉資料以形成聯合對數機率的 部分應用 的結果。
# Define the joint_log_prob function, and our unnormalized posterior.
def joint_log_prob(w, x, y):
# Our model in maths is
# w ~ MVN([0, 0, 0], diag([1, 1, 1]))
# y_i ~ Normal(w @ x_i, noise_scale), i=1..N
rv_w = tfd.MultivariateNormalDiag(
loc=np.zeros(num_features + 1),
scale_diag=np.ones(num_features + 1))
rv_y = tfd.Normal(f(x, w), noise_scale)
return (rv_w.log_prob(w) +
tf.reduce_sum(rv_y.log_prob(y), axis=-1))
# Create our unnormalized target density by currying x and y from the joint.
def unnormalized_posterior(w):
return joint_log_prob(w, xs, ys)
建構 HMC TransitionKernel 並呼叫 sample_chain
# Create an HMC TransitionKernel
hmc_kernel = tfp.mcmc.HamiltonianMonteCarlo(
target_log_prob_fn=unnormalized_posterior,
step_size=np.float64(.1),
num_leapfrog_steps=2)
# We wrap sample_chain in tf.function, telling TF to precompile a reusable
# computation graph, which will dramatically improve performance.
@tf.function
def run_chain(initial_state, num_results=1000, num_burnin_steps=500):
return tfp.mcmc.sample_chain(
num_results=num_results,
num_burnin_steps=num_burnin_steps,
current_state=initial_state,
kernel=hmc_kernel,
trace_fn=lambda current_state, kernel_results: kernel_results)
initial_state = np.zeros(num_features + 1)
samples, kernel_results = run_chain(initial_state)
print("Acceptance rate:", kernel_results.is_accepted.numpy().mean())
Acceptance rate: 0.915
這不太好!我們希望接受率更接近 0.65。
(請參閱 「各種 Metropolis-Hastings 演算法的最佳縮放比例」,Roberts & Rosenthal,2001)
自適應步長
我們可以將我們的 HMC TransitionKernel 包裝在 SimpleStepSizeAdaptation「中繼核心」中,這將應用一些 (相當簡單的啟發式) 邏輯,以便在預熱期間調整 HMC 步長。我們分配 80% 的預熱時間用於調整步長,然後讓剩餘的 20% 僅用於混合。
# Apply a simple step size adaptation during burnin
@tf.function
def run_chain(initial_state, num_results=1000, num_burnin_steps=500):
adaptive_kernel = tfp.mcmc.SimpleStepSizeAdaptation(
hmc_kernel,
num_adaptation_steps=int(.8 * num_burnin_steps),
target_accept_prob=np.float64(.65))
return tfp.mcmc.sample_chain(
num_results=num_results,
num_burnin_steps=num_burnin_steps,
current_state=initial_state,
kernel=adaptive_kernel,
trace_fn=lambda cs, kr: kr)
samples, kernel_results = run_chain(
initial_state=np.zeros(num_features+1))
print("Acceptance rate:", kernel_results.inner_results.is_accepted.numpy().mean())
Acceptance rate: 0.634
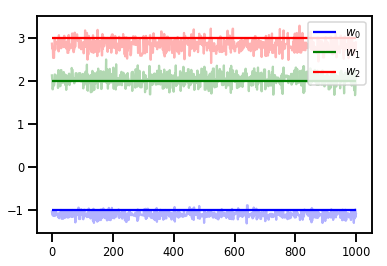
# Trace plots
colors = ['b', 'g', 'r']
for i in range(3):
plt.plot(samples[:, i], c=colors[i], alpha=.3)
plt.hlines(true_w[i], 0, 1000, zorder=4, color=colors[i], label="$w_{}$".format(i))
plt.legend(loc='upper right')
plt.show()
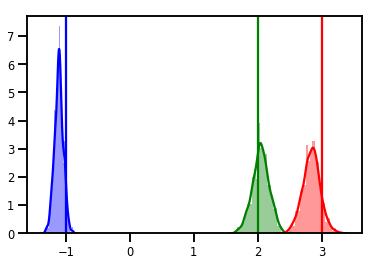
# Histogram of samples
for i in range(3):
sns.distplot(samples[:, i], color=colors[i])
ymax = plt.ylim()[1]
for i in range(3):
plt.vlines(true_w[i], 0, ymax, color=colors[i])
plt.ylim(0, ymax)
plt.show()
診斷
追蹤圖很棒,但診斷更好!
首先,我們需要執行多個鏈。這就像給定一批 initial_state 張量一樣簡單。
# Instead of a single set of initial w's, we create a batch of 8.
num_chains = 8
initial_state = np.zeros([num_chains, num_features + 1])
chains, kernel_results = run_chain(initial_state)
r_hat = tfp.mcmc.potential_scale_reduction(chains)
print("Acceptance rate:", kernel_results.inner_results.is_accepted.numpy().mean())
print("R-hat diagnostic (per latent variable):", r_hat.numpy())
Acceptance rate: 0.59175 R-hat diagnostic (per latent variable): [0.99998395 0.99932185 0.9997064 ]
取樣雜訊尺度
# Define the joint_log_prob function, and our unnormalized posterior.
def joint_log_prob(w, sigma, x, y):
# Our model in maths is
# w ~ MVN([0, 0, 0], diag([1, 1, 1]))
# y_i ~ Normal(w @ x_i, noise_scale), i=1..N
rv_w = tfd.MultivariateNormalDiag(
loc=np.zeros(num_features + 1),
scale_diag=np.ones(num_features + 1))
rv_sigma = tfd.LogNormal(np.float64(1.), np.float64(5.))
rv_y = tfd.Normal(f(x, w), sigma[..., np.newaxis])
return (rv_w.log_prob(w) +
rv_sigma.log_prob(sigma) +
tf.reduce_sum(rv_y.log_prob(y), axis=-1))
# Create our unnormalized target density by currying x and y from the joint.
def unnormalized_posterior(w, sigma):
return joint_log_prob(w, sigma, xs, ys)
# Create an HMC TransitionKernel
hmc_kernel = tfp.mcmc.HamiltonianMonteCarlo(
target_log_prob_fn=unnormalized_posterior,
step_size=np.float64(.1),
num_leapfrog_steps=4)
# Create a TransformedTransitionKernl
transformed_kernel = tfp.mcmc.TransformedTransitionKernel(
inner_kernel=hmc_kernel,
bijector=[tfb.Identity(), # w
tfb.Invert(tfb.Softplus())]) # sigma
# Apply a simple step size adaptation during burnin
@tf.function
def run_chain(initial_state, num_results=1000, num_burnin_steps=500):
adaptive_kernel = tfp.mcmc.SimpleStepSizeAdaptation(
transformed_kernel,
num_adaptation_steps=int(.8 * num_burnin_steps),
target_accept_prob=np.float64(.75))
return tfp.mcmc.sample_chain(
num_results=num_results,
num_burnin_steps=num_burnin_steps,
current_state=initial_state,
kernel=adaptive_kernel,
seed=(0, 1),
trace_fn=lambda cs, kr: kr)
# Instead of a single set of initial w's, we create a batch of 8.
num_chains = 8
initial_state = [np.zeros([num_chains, num_features + 1]),
.54 * np.ones([num_chains], dtype=np.float64)]
chains, kernel_results = run_chain(initial_state)
r_hat = tfp.mcmc.potential_scale_reduction(chains)
print("Acceptance rate:", kernel_results.inner_results.inner_results.is_accepted.numpy().mean())
print("R-hat diagnostic (per w variable):", r_hat[0].numpy())
print("R-hat diagnostic (sigma):", r_hat[1].numpy())
Acceptance rate: 0.715875 R-hat diagnostic (per w variable): [1.0000073 1.00458208 1.00450512] R-hat diagnostic (sigma): 1.0092056996149859
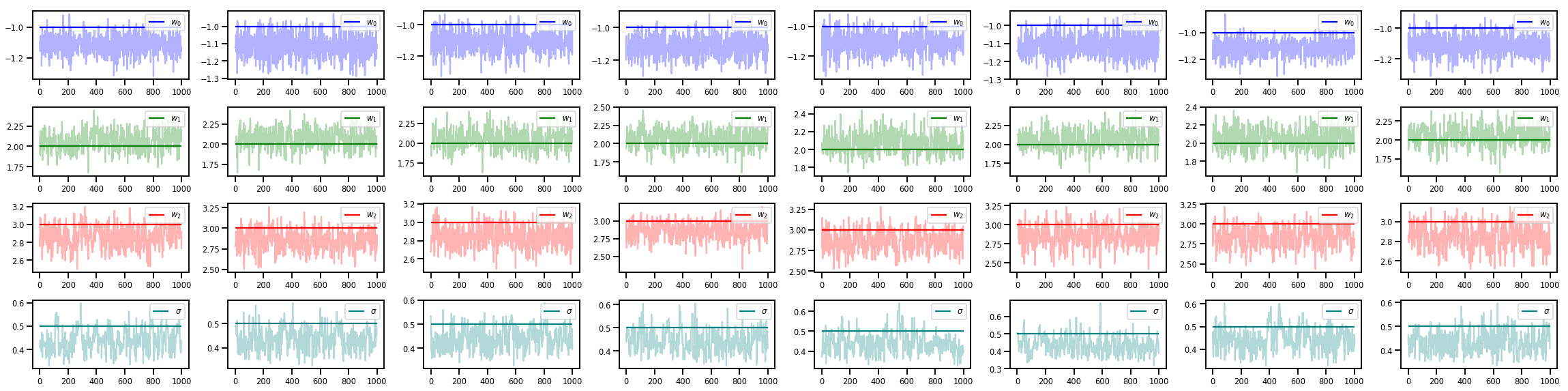
w_chains, sigma_chains = chains
# Trace plots of w (one of 8 chains)
colors = ['b', 'g', 'r', 'teal']
fig, axes = plt.subplots(4, num_chains, figsize=(4 * num_chains, 8))
for j in range(num_chains):
for i in range(3):
ax = axes[i][j]
ax.plot(w_chains[:, j, i], c=colors[i], alpha=.3)
ax.hlines(true_w[i], 0, 1000, zorder=4, color=colors[i], label="$w_{}$".format(i))
ax.legend(loc='upper right')
ax = axes[3][j]
ax.plot(sigma_chains[:, j], alpha=.3, c=colors[3])
ax.hlines(noise_scale, 0, 1000, zorder=4, color=colors[3], label=r"$\sigma$".format(i))
ax.legend(loc='upper right')
fig.tight_layout()
plt.show()
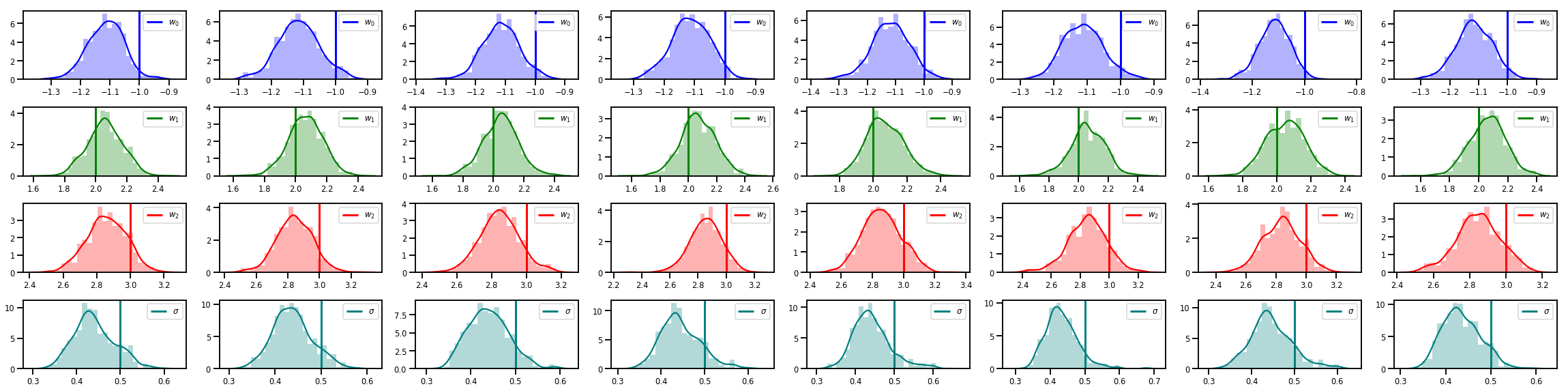
# Histogram of samples of w
fig, axes = plt.subplots(4, num_chains, figsize=(4 * num_chains, 8))
for j in range(num_chains):
for i in range(3):
ax = axes[i][j]
sns.distplot(w_chains[:, j, i], color=colors[i], norm_hist=True, ax=ax, hist_kws={'alpha': .3})
for i in range(3):
ax = axes[i][j]
ymax = ax.get_ylim()[1]
ax.vlines(true_w[i], 0, ymax, color=colors[i], label="$w_{}$".format(i), linewidth=3)
ax.set_ylim(0, ymax)
ax.legend(loc='upper right')
ax = axes[3][j]
sns.distplot(sigma_chains[:, j], color=colors[3], norm_hist=True, ax=ax, hist_kws={'alpha': .3})
ymax = ax.get_ylim()[1]
ax.vlines(noise_scale, 0, ymax, color=colors[3], label=r"$\sigma$".format(i), linewidth=3)
ax.set_ylim(0, ymax)
ax.legend(loc='upper right')
fig.tight_layout()
plt.show()
還有更多!
查看這些酷炫的部落格文章和範例
更多範例和筆記本,請參閱我們的 GitHub 此處!
